MSc in Bioinformatics – Computer Science for Biology and Health
University of Rennes 1 | 2024 – 2025

Overview
To develop additional research competencies and bridge the gap between experimental biology and computational data analysis, I pursued a specialized MSc in Bioinformatics applied to Biomedical and Health Sciences. This complementary training not only equipped me with the advanced bioinformatics skills necessary to independently conduct modern biological research—where high-throughput data generation and computational analysis have become essential—but also allowed me to expand my scope toward human biology and medicine, a long-standing interest of mine.
Through the CoCoBi minor (Complementary Skills in Bioinformatics), I gained direct admission to this intensive M2 program at the University of Rennes 1.
Core Competencies
Programming & Development
- Proficient in Python and Bash for data analysis and automation
- Version control and collaborative development with Git
- Workflow automation and reproducibility with Nextflow
Environment & Infrastructure Management
- Package and environment management with Conda and uv
- Containerization with Docker and Singularity for reproducible deployments
- High-Performance Computing (HPC) platforms for large-scale data processing
Reproducible Research Practices
- Implementation of FAIR principles (Findable, Accessible, Interoperable, Reusable)
- Documentation with Quarto for professional reports and presentations
- Interactive data analysis and exploration using Jupyter and Marimo notebooks
- Project organization and data management best practices (AGILE methodology)
Statistics & Machine Learning
Application of advanced statistical methods and machine learning algorithms to biological data:
- Regression models (linear, logistic, generalized linear models)
- Decision Trees and Random Forests
- Neural Networks and Deep Learning
- Support Vector Machines (SVM)
- Principal Component Analysis (PCA) and dimensionality reduction
- Clustering methods (k-means, hierarchical clustering)
Omics Data Analysis
Analysis, interpretation, and visualization of massive omics datasets with applications in diagnostics, biomedical research, and fundamental biology:
- Metagenomic Analysis in Ecology
- Molecular Evolution and Phylogenomics
- Graph Theory and Network Analysis
- Genome Assembly and Annotation
- Functional Genomics: bulk and scRNA-seq, ATAC-seq on human data
Key Academic Projects
Throughout this Master’s program, I completed several hands-on bioinformatics projects that reinforced theoretical concepts and developed practical computational skills. Below are representative projects demonstrating my ability to design pipelines, analyze complex datasets, and communicate technical findings.
Professional Research Experience
As part of this Master’s program, I completed a 6-month research internship to apply these competencies in a real-world research environment and further develop my expertise in computational biology.
MSc Research Intern – Bioinformatics & Microbial Ecology
Feb. – July 2025 | UMR 6553 ECOBIO, Rennes
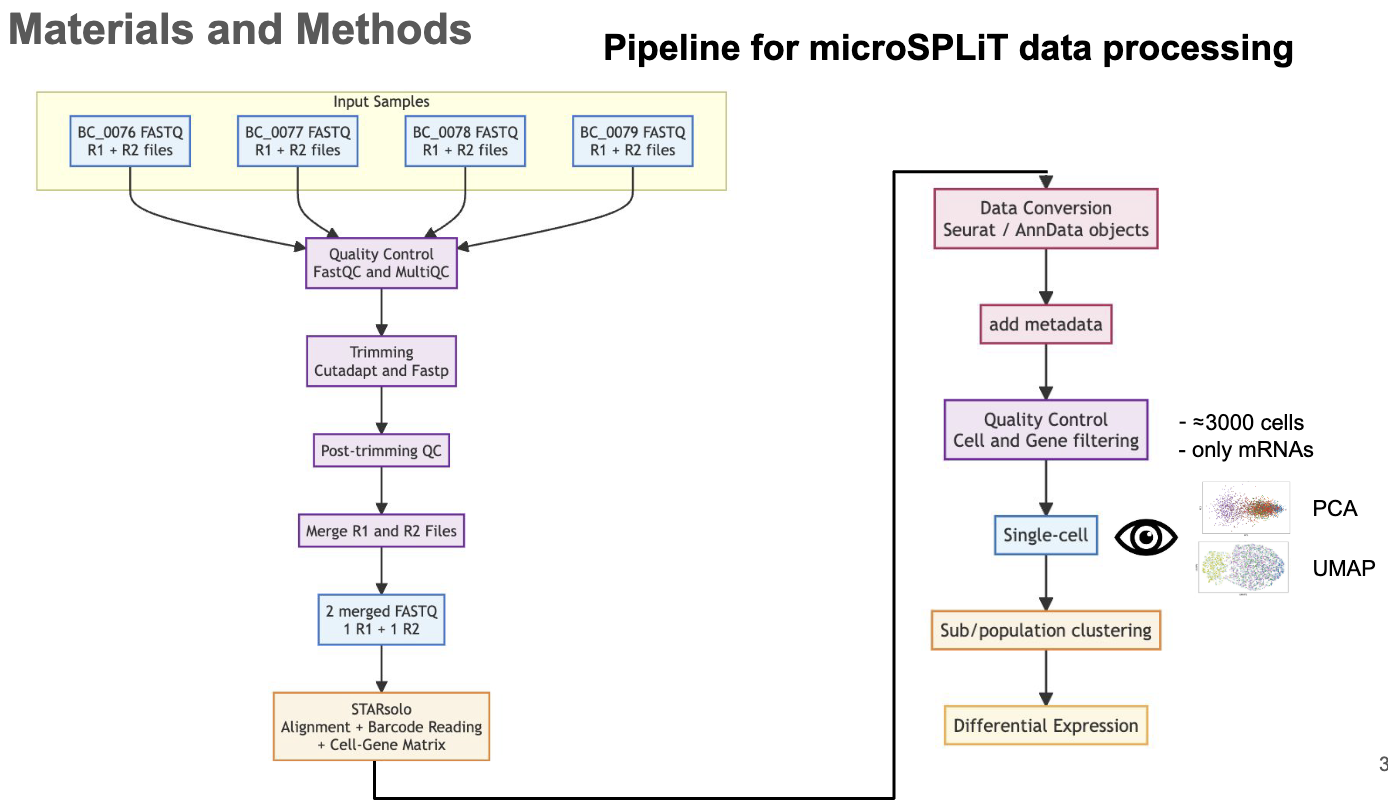
Performed scRNA-seq analyses to study division of labor within genetically identical bacterial populations (Pseudomonas brassicacearum) as part of the ANR DivIDE project.
Supervised by Philippe Vandenkoornhuyse & Solène Mauger-Franklin


